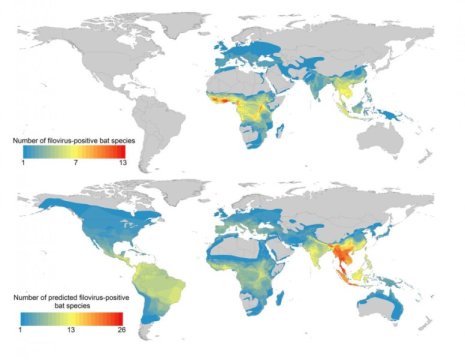
An international team of ecologists has identified the bat species with the greatest potential to harbor filoviruses — a family that includes Ebola virus. Writing in PLOS Neglected Tropical Diseases, they suggest that areas where many of these species overlap, notably in Southeast Asia, should be targeted for disease surveillance and virus discovery efforts.
“Using machine learning methods developed for artificial intelligence, we were able to bring together data from ecology, biogeography and public health to identify bat species with a high probability of harboring Ebola and other filoviruses,” said the paper’s lead author Barbara Han of the Cary Institute of Ecosystem Studies. “Understanding which species carry these viruses, and where they are located, is essential to preventing future spillovers.”
Filoviruses, which originate in animals and can be transmitted to humans, pose a serious threat to human health and to primate conservation. Ebola virus disease has a 50 percent human fatality rate, according to the World Health Organization, and among great apes like gorillas and chimpanzees that rate is even higher.
Identifying which animals serve as Ebola virus reservoirs — sources from which it can spread to humans and other susceptible animals — is critical for preventing future outbreaks. This has proven difficult in part because of the enormous geographic range of the virus across equatorial Africa and the sheer number of potential reservoir species within that range. There is mounting evidence, however, that certain bats are the likeliest culprits.
Han and colleagues from the University of Georgia and Massey University in New Zealand set out to develop a “trait profile” of filovirus-susceptible bats. Looking at more than 50 biological and ecological traits of the 21 bat species known to harbor filoviruses, they found several that distinguish them from other bats with 87 percent accuracy. These include earlier maturity, more frequent litters, and pups that are larger at birth. Their geographic ranges are also larger, and overlap with more mammal species, than those of other bats.
They used machine learning — complex computer programs that can analyze enormous amounts of data and find hidden patterns — to compare all 1,116 of the world’s bat species to the profile. This allowed them to determine which other species share those traits and are therefore potential filovirus hosts. Finally, they mapped the geographic ranges of all the species that fit the trait profile to pinpoint potential filovirus hotspots.
Their results contained some surprises.
While many of the species that most closely fit the profile are found in sub-Saharan Africa, the top 10 percent of likeliest hosts are much more widely distributed than the researchers expected. Species range across Southeast Asia and Central and South America. Several hotspots, where a number of potential reservoir species overlap, are outside Africa, most notably in parts of Thailand, Burma, Malaysia, Vietnam and Northeastern India.
The findings provide information that could be used to target surveillance more efficiently by focusing on the likeliest filovirus carriers, many of which have never been tested.
Senior author John Drake, director of the new UGA Center for the Ecology of Infectious Diseases at the Odum School of Ecology, said that the results also highlight the importance of the multidisciplinary approach that is the center’s hallmark.
“One thing the center aims to do is to perform synthetic (multi-disciplinary) research that reaches from basic science all the way out to wildlife management, public health and clinical practice,” he said. “This work exemplifies how the center supports such endeavors by bringing together subject matter experts (in this case David Hayman of Massey University) with ‘big picture’ scientific visionaries like Barbara Han via the methodology of modern data science (UGA’s Drake and J.P Schmidt).”
In addition, UGA undergraduate Laura Alexander, now a doctoral student at the University of California, Berkeley, reviewed the primary literature to compile a comprehensive picture of what was and wasn’t known about the subject area, and UGA doctoral student Sarah Bowden — now a postdoctoral scientist at the Cary Institute — developed the maps.
The findings have raised questions for future research that will require a similar multidisciplinary approach.
“We suspect there may be other filoviruses waiting to be found,” Drake said. “An outstanding question for future work is to investigate why there are so few filovirus spillover events reported for humans and wildlife in Southeast Asia compared to equatorial Africa.”
Support for the research was provided by the National Institutes of Health RAPIDD program and NIH National Institute of General Medical Sciences under Award Number U01GM110744.
Story Source:
The above post is reprinted from materials provided byUniversity of Georgia. The original item was written by Beth Gavrilles. Note: Materials may be edited for content and length.
Journal Reference:
- Barbara A. Han, John Paul Schmidt, Laura W. Alexander, Sarah E. Bowden, David T. S. Hayman, John M. Drake.Undiscovered Bat Hosts of Filoviruses. PLOS Neglected Tropical Diseases, 2016; 10 (7): e0004815 DOI:10.1371/journal.pntd.0004815