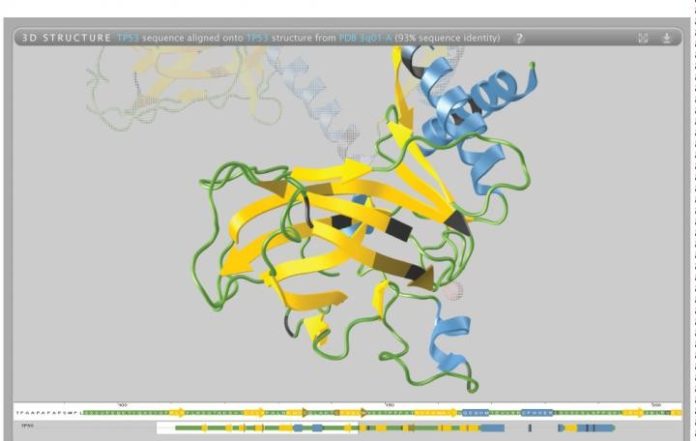
Life scientists now have access to a publicly available web resource that streamlines and simplifies the process of gleaning insight from 3D protein structures. Known as Aquaria, the powerful tool is announced today in Nature Methods.
The Aquaria project was led by Dr Seán O’Donoghue, from The Garvan Institute of Medical Research and CSIRO, in collaboration with Dr Andrea Schafferhans from the Technical University of Munich. The project started in 2009 and involved an international team of around a dozen programmers and bioinformatics experts.
Aquaria is built upon the Protein Data Bank, which contains just over 100,000 protein structures.
“The Protein Data Bank is a fantastic resource containing a wealth of detail about the molecular processes of life, but we were aware that few biologists take full advantage of it,” said O’Donoghue.
“So we created Aquaria to make this valuable information more accessible and easier to use for discovery purposes.
“What we’ve done is to layer in a lot of extra useful information. For example, we’ve added protein sequences that do not yet have a structure — but are similar to something in the Protein Data Bank. That meant we first had to find all these similarities.
“So we took over 500,000 protein sequences and compared every one of them with the 100,000 known protein structures — and that has given us around 46 million computer models.
“Aquaria is fast, it comes with an easy-to-use interface and contains twice as many models as all other similar resources combined. It also allows users to view additional information — such as genetic differences between individuals — mapped onto 3D structures.
“For example, you can add Single Nucleotide Polymorphisms, or ‘SNPs’, that cause protein changes, then visualise exactly where those changes occur in the protein structure. This provides valuable insight into why proteins sometimes completely change their function as a result of one small change in the DNA code.
“You can then ask interesting questions like ‘Does this set of SNPs cluster in 3D?’ and the answers to such questions can set new research directions.”
Aquaria will be useful to a broad range of life scientists, from medical researchers — at institutes like Garvan — to scientists studying agriculture, biosecurity, ecology and nutrition at institutes like CSIRO.
Aquaria’s flexibility and extensibility allows information to be combined in completely new ways — quickly and easily. All a scientist needs to do is enter the name of their favourite protein, and then navigate a brave new world of possibility.
Story Source:
The above story is based on materials provided by Garvan Institute of Medical Research. Note: Materials may be edited for content and length.
Journal Reference:
- Seán I O’Donoghue, Kenneth S Sabir, Maria Kalemanov, Christian Stolte, Benjamin Wellmann, Vivian Ho, Manfred Roos, Nelson Perdigão, Fabian A Buske, Julian Heinrich, Burkhard Rost, Andrea Schafferhans. Aquaria: simplifying discovery and insight from protein structures. Nature Methods, 2015; 12 (2): 98 DOI: 10.1038/nmeth.3258