Humans don’t like being alone, and their genes are no different. Together we are stronger, and the two versions of a gene — one from each parent — need each other. Scientists at the Max Planck Institute for Molecular Genetics in Berlin have analysed the genetic makeup of several hundred people and decoded the genetic information on the two sets of chromosomes separately. In this relatively small group alone they found millions of different gene forms. The results also show that genetic mutations do not occur randomly in the two parental chromosome sets and that they are distributed in the same ratio in everyone.
In 2001 scientists announced the successful decoding of the first human genome. Since then, thousands more have been sequenced. The price of a genetic analysis will soon fall below the 1,000 dollar mark. Given this rapid pace of development, it’s easy to forget that the technology used only reads a mixed product of genetic information. The analytical methods commonly employed do not take into account the fact that every person has two sets of genetic material. “So they are ignoring an essential property of the human genome. However, it’s important to know, for example, how mutations are distributed between the two chromosome sets,” says Margret Hoehe from the Max Planck Institute for Molecular Genetics, who carried out the study.
Hoehe and her team have developed molecular genetic and bioinformatic methods that make it possible to sequence the two sets of chromosomes in a human separately. The researchers decoded the maternal and paternal parts of the genome in 14 people and supplemented their analysis with the genetic material of 372 Europeans from the 1000 Genomes Project. “Fourteen people may not sound like a lot, but given the technical challenge, it is an unprecedented achievement,” says Hoehe.
The results show that most genes can occur in many different forms within a population: On average, about 250 different forms of each gene exist. The researchers found around four million different gene forms just in the 400 or so genomes they analysed. This figure is certain to increase as more human genomes are examined. More than 85 percent of all genes have no predominant form which occurs in more than half of all individuals. This enormous diversity means that over half of all genes in an individual, around 9,000 of 17,500, occur uniquely in that one person — and are therefore individual in the truest sense of the word.
The gene, as we imagined it, exists only in exceptional cases. “We need to fundamentally rethink the view of genes that every schoolchild has learned since Gregor Mendel’s time. Moreover, the conventional view of individual mutations is no longer adequate. Instead, we have to consider the two gene forms and their combination of variants,” Hoehe explains. When analysing genomes, scientists should therefore examine each parental gene form separately, as well as the effects of both forms as a pair.
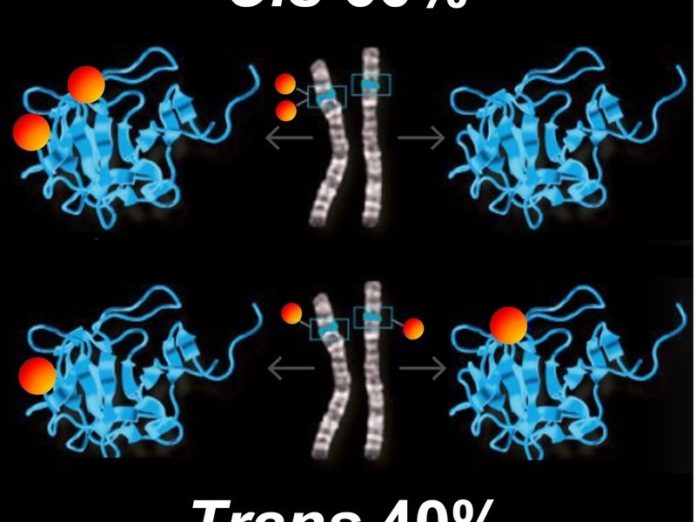
According to the researchers, mutations of genes are not randomly distributed between the parental chromosomes. They found that 60 percent of mutations affect the same chromosome set and 40 percent both sets. Scientists refer to these as cis and trans mutations, respectively. Evidently, an organism must have more cis mutations, where the second gene form remains intact. “It’s amazing how precisely the 60:40 ratio is maintained. It occurs in the genome of every individual — almost like a magic formula,” says Hoehe. The 60:40 distribution ratio appears to be essential for survival. “This formula may help us to understand how gene variability occurs and how it affects gene function.”
Some of the many variants that alter the genome also have an effect at the protein level. The researchers have now identified a set of 4,000 genes that are altered by mutations so that their proteins occur especially frequently in two different forms in humans. These genes mainly control signal transmission between cells, the immune system and gene activity. This dual gene and protein arrangement has the advantage that it allows the activity of genes to be more flexibly adjusted and altered. By using the more favourable variant, the body is better able to adapt to changes in its own processes and to environmental conditions. If the duality of genes goes awry and the wrong protein form is used, this can trigger pathogenic mechanisms. This is probably why those 4,000 genes include many disease genes.
These findings will change the interpretation of genetic analyses and the prediction of diseases. Moreover, individualised medicine cannot ignore the “dual nature” of human genomes. “Our investigations at the protein level have shown that 96 percent of all genes have at least 5 to 20 different protein forms. This results in tremendous individual diversity in possible interactions between genes, and shows how daunting the challenge is to develop individually tailored therapies,” says Hoehe.
So far, researchers have estimated the risk of disease only by the presence or absence of mutations. However, there is evidence that in cancer, for example, the severity and course of the disease is determined by the wrong distribution of a mutation. The location of mutations therefore needs to be considered in the diagnosis, prediction and prevention of diseases in future.
Story Source:
The above story is based on materials provided by Max-Planck-Gesellschaft. Note: Materials may be edited for content and length.
Journal Reference:
- Margret R. Hoehe, George M. Church, Hans Lehrach, Thomas Kroslak, Stefanie Palczewski, Katja Nowick, Sabrina Schulz, Eun-Kyung Suk, Thomas Huebsch. Multiple haplotype-resolved genomes reveal population patterns of gene and protein diplotypes. Nature Communications, 2014; 5: 5569 DOI: 10.1038/ncomms6569