Scientists looking to understand — and potentially thwart — the influenza virus now have a much more encompassing view, thanks to the first complete structure of one of the flu virus’ key machines. The structure, obtained by scientists at EMBL Grenoble, allows researchers to finally understand how the machine works as a whole, and could prove instrumental in designing new drugs to treat serious flu infections and combat flu pandemics.
If you planned to sabotage a factory, a recon trip through the premises would probably be much more useful than just peeping in at the windows. Scientists looking to understand — and potentially thwart — the influenza virus have now gone from a similar window-based view to the full factory tour, thanks to the first complete structure of one of the flu virus’ key machines. The structure, obtained by scientists at the European Molecular Biology Laboratory (EMBL) in Grenoble, France, allows researchers to finally understand how the machine works as a whole. Published in two papers in Nature, the work could prove instrumental in designing new drugs to treat serious flu infections and combat flu pandemics.
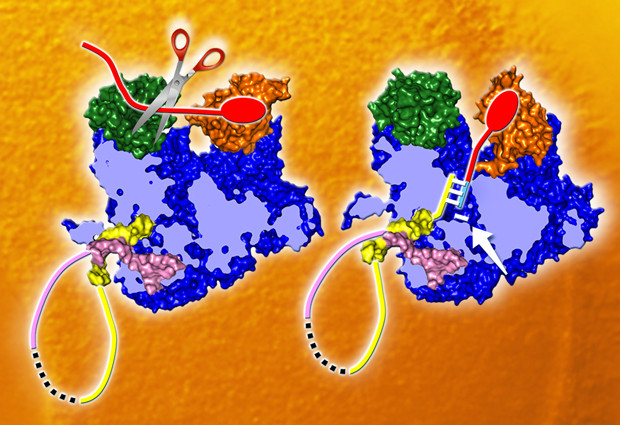
The machine in question, the influenza virus polymerase, carries out two vital tasks for the virus. It makes copies of the virus’ genetic material — the viral RNA — to package into new viruses that can infect other cells; and it reads out the instructions in that genetic material to make viral messenger RNA, which directs the infected cell to produce the proteins the virus needs. Scientists — including Cusack and collaborators — had been able to determine the structure of several parts of the polymerase in the past. But how those parts came together to function as a whole, and how viral RNA being fed in to the polymerase could be treated in two different ways remained a mystery.
“The flu polymerase was discovered 40 years ago, so there are hundreds of papers out there trying to fathom how it works. But only now that we have the complete structure can we really begin to understand it,” says Stephen Cusack, head of EMBL Grenoble, who led the work.
Using X-ray crystallography, performed at the European Synchrotron Radiation Facility (ESRF) in Grenoble, Cusack and colleagues were able to determine the atomic structure of the whole polymerase from two strains of influenza: influenza B, one of the strains that cause seasonal flu in humans, but which evolves slowly and therefore isn’t considered a pandemic threat; and the strain of influenza A — the fast-evolving strain that affects humans, birds and other animals and can cause pandemics — that infects bats.
“The high-intensity X-ray beamlines at the ESRF, equipped with state-of-the-art Dectris detectors, were crucial for getting high quality crystallographic data from the weakly diffracting and radiation sensitive crystals of the large polymerase complex,” says Cusack. “We couldn’t have got the data at such a good resolution without them.”
The structures reveal how the polymerase specifically recognises and binds to the viral RNA, rather than just any available RNA, and how that binding activates the machine. They also show that the three component proteins that make up the polymerase are very intertwined, which explains why it has been very difficult to piece together how this machine works based on structures of individual parts.
Although the structures of both viruses’ polymerases were very similar, the scientists found one key difference, which showed that one part of the machine can swivel around to a large degree. That ability to swivel explains exactly how the polymerase uses host cell RNA to kick-start the production of viral proteins. The swivelling component takes the necessary piece of host cell RNA and directs it into a slot leading to the machine’s heart, where it triggers the production of viral messenger RNA.
Now that they know exactly where each atom fits in this key viral machine, researchers aiming to design drugs to stop influenza in its tracks have a much wider range of potential targets at their disposal — like would-be saboteurs who gain access to the whole production plant instead of just sneaking looks through the windows. And because this is such a fundamental piece of the viral machinery, not only are the versions in the different influenza strains very similar to each other, but they also hold many similarities to their counterparts in related viruses such as lassa, hanta, rabies or ebola.
The EMBL scientists aim to explore the new insights this structure provides for drug design, as well as continuing to try to determine the structure of the human version of influenza A, because although the bat version is close enough that it already provides remarkable insights, ultimately fine-tuning drugs for treating people would benefit from/require knowledge of the version of the virus that infects humans. And, since this viral machine has to be flexible and change shape to carry out its different tasks, Cusack and colleagues also want to get further snapshots of the polymerase in different states.
“This doesn’t mean we now have all the answers,” says Cusack, “In fact, we have as many new questions as answers, but at least now we have a solid basis on which to probe further.”
Story Source:
The above story is based on materials provided by European Molecular Biology Laboratory (EMBL). Note: Materials may be edited for content and length.
Journal References:
- Alexander Pflug, Delphine Guilligay, Stefan Reich, Stephen Cusack. Structure of influenza A polymerase bound to the viral RNA promoter. Nature, 2014; DOI: 10.1038/nature14008
- Stefan Reich, Delphine Guilligay, Alexander Pflug, Hélène Malet, Imre Berger, Thibaut Crépin, Darren Hart, Thomas Lunardi, Max Nanao, Rob W. H. Ruigrok, Stephen Cusack. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature, 2014; DOI: 10.1038/nature14009