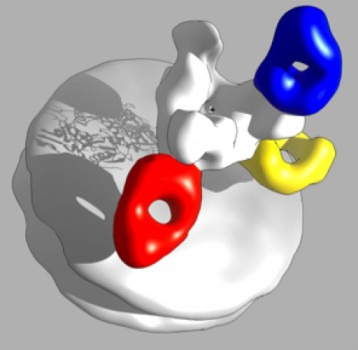
Scientists at The Scripps Research Institute (TSRI) have identified weak spots on the surface of Ebola virus that are targeted by the antibodies in ZMapp, the experimental drug cocktail administered to several patients during the recent Ebola outbreak.
The study, led by TSRI structural biologists Andrew Ward and Erica Ollmann Saphire and published online ahead of print this week by the journal Proceedings of the National Academy of Sciences, provides a revealing 3-D picture of how the ZMapp antibodies bind to Ebola virus.
“The structural images of Ebola virus are like enemy reconnaissance,” said Saphire. “They tell us exactly where to target antibodies or drugs.”
Ward said, “Now that we know how ZMapp targets Ebola, we can compare all newly discovered anti-Ebola antibodies as we try to formulate an even better immunotherapeutic cocktail.”
How Antibodies Fight Back
ZMapp, developed by San Diego-based Mapp Biopharmaceutical, was used in August to treat several patients in the ongoing Ebola virus outbreak. Although five of the seven patients who received ZMapp survived, researchers cannot yet say for sure whether ZMapp made a difference in their recoveries.
The new study explains why ZMapp could have been effective. Using an imaging technique called electron microscopy, researchers found that two of the ZMapp antibodies bind near the base of virus, appearing to prevent the virus from entering cells. A third antibody binds near the top of the virus, possibly acting as a beacon to call the body’s immune system to the site of infection.
The new picture of ZMapp reveals the two antibodies that bind near the base of the virus seem to be competing for the same site. While this appears to be a particularly vulnerable spot on Ebola virus’s surface as identified in previous studies, one question now is whether future cocktails should continue to use two antibodies to target this site or try to attack the virus from a third angle.
“This information helps guide decisions about how to formulate these life-saving therapies,” said C. Daniel Murin, a graduate student in the labs of Ward and Saphire and first author of the new study. “Instead of including two different antibodies that do the same thing, why not use twice as much of the more effective one instead? Or include a third antibody against a different site to stop the virus a third way?”
Luckily, while the Ebola virus has undergone more than 300 genetic changes in the current outbreak (according to research published in the journal Science in August), the new study indicates the sites where the ZMapp antibodies bind have been unaffected so far.
A Global Search for Treatments
The new research is part of the National Institutes of Health-funded Viral Hemorrhagic Fever Immunotherapeutic Consortium, which is testing antibodies from 25 laboratories around the world with the goal of developing the best cocktail for neutralizing Ebola virus and other closely related hemorrhagic fever viruses.
The next step for the consortium is to study the new antibodies from human survivors of the current outbreak. Saphire, who leads the consortium, hopes the group can also develop a back-up cocktail in case the virus mutates and becomes resistant to treatment.
ZMapp is expected to go into clinical trials in early 2015. The antibodies in the cocktail were originally isolated by the Public Health Agency of Canada and the U.S. Army Medical Research Institute of Infectious Disease.
In addition to Ward, Saphire and Murin, the authors of the paper “Structures of protective antibodies reveal sites of vulnerability on Ebola virus” were Marnie L. Fusco and Zachary A. Bornholdt of TSRI; Xiangguo Qiu of the Public Health Agency of Canada; Gene G. Olinger of the National Institutes of Health; Larry Zeitlin of Mapp Biopharmaceutical; and Gary P. Kobinger of the Public Health Agency of Canada and the University of Manitoba.
Funding for the research was provided by the National Institutes of Health (R01AI067927 and U19AI109762), the National Institute of General Medical Sciences (GM103310), the National Science Foundation, the Ray Thomas Edwards Foundation and the Burroughs Welcome Fund.
Story Source:
The above story is based on materials provided by Scripps Research Institute. Note: Materials may be edited for content and length.
Journal Reference:
- Charles D. Murin, Marnie L. Fusco, Zachary A. Bornholdt, Xiangguo Qiu, Gene G. Olinger, Larry Zeitlin, Gary P. Kobinger, Andrew B. Ward, and Erica Ollmann Saphire. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. PNAS, November 17, 2014 DOI: 10.1073/pnas.1414164111