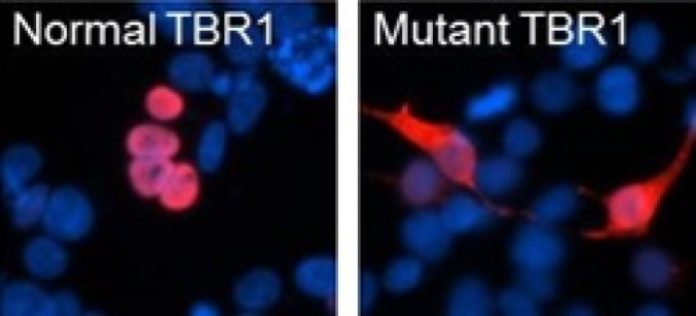
Spontaneous mutations in the brain gene TBR1 disrupt the function of the encoded protein in children with severe autism. In addition, there is a direct link between TBR1 and FOXP2, a well-known language-related protein. These are the main findings of Pelagia Deriziotis and colleagues at the Nijmegen Max Planck Institute for Psycholinguistics in an article published by Nature Communications on September 18.
Autism is a disorder of brain development which leads to difficulties with social interaction and communication. Disorders such as autism are often caused by genetic mutations, which can change the shape of protein molecules and stop them from working properly during brain development. In some individuals with autism, inherited genetic variants put them at risk. But research in recent years has shown that severe cases of autism can result from new mutations occurring in the sperm or egg — these genetic variants are found in a child, but not in his or her parents, and are known as de novo mutations. Scientists have sequenced the DNA code of thousands of unrelated children with severe autism and found that a handful of genes are hit by independent de novo mutations in more than one child. One of the most interesting of these genes is TBR1, a key gene in brain development.
Strong impact on protein function In their study, Pelagia Deriziotis and colleagues from the MPI’s Language and Genetics Department and the University of Washington investigated the effects of autism risk mutations on TBR1 protein function. They used several cutting-edge techniques to examine how these mutations affect the way the TBR1 protein works, using human cells grown in the laboratory. ‘We directly compared de novo and inherited mutations, and found that the de novo mutations had much more dramatic effects on TBR1 protein function’, says Dr. Deriziotis, ‘This is a really striking confirmation of the strong impact that de novo mutations can have on early brain development’.
Social network for proteins Since the human brain depends on many different genes and proteins working together, the researchers were interested in identifying proteins that interact with TBR1. They discovered that TBR1 directly interacts with FOXP2, an important protein in speech and language disorders, and that pathogenic mutations affecting either of these proteins abolish the mutual interaction. ‘Think of it as a social network for proteins’, says Dr. Deriziotis, ‘There were initial clues that TBR1 might be “friends” with FOXP2. This was intriguing because FOXP2 is one of the few proteins to have been clearly implicated in speech and language disorders. The common molecular pathway we found is very interesting.’
According to senior author Simon Fisher, Professor of Language and Genetics at Radboud University and director at the MPI: ‘It is very exciting to uncover these fascinating molecular links between different disorders that affect language. By coupling data from genome screening with functional analysis in the lab, we are starting to build up a picture of the neurogenetic pathways that contribute to fundamental human traits.’
Story Source:
The above story is based on materials provided by Radboud University Nijmegen. Note: Materials may be edited for content and length.
Journal Reference:
- Pelagia Deriziotis, Brian J. O’Roak, Sarah A. Graham, Sara B. Estruch, Danai Dimitropoulou, Raphael A. Bernier, Jennifer Gerdts, Jay Shendure, Evan E. Eichler, Simon E. Fisher. De novo TBR1 mutations in sporadic autism disrupt protein functions. Nature Communications, 2014; 5: 4954 DOI: 10.1038/ncomms5954