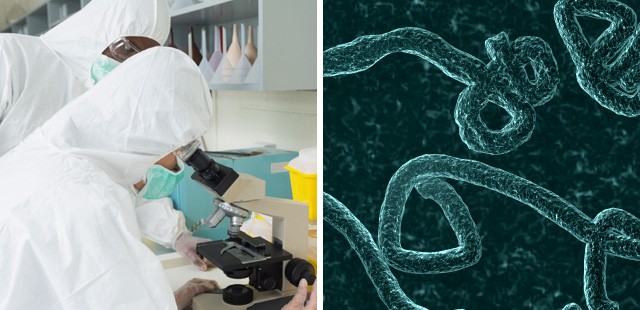
For the first time, scientists have been able to follow the spread of an Ebola outbreak almost in real time, by sequencing the virus’ genome from people in Sierra Leone. The findings, published Thursday in the journal Science, offer new insights into how the outbreak started in West Africa and how rapidly the virus is mutating.
The current Ebola outbreak, which began back in late winter/early spring and is still picking up speed, is somewhat of an anomaly. Previous outbreaks have been confined to eastern and southern regions of Africa, primarily affecting countries such as the Democratic Republic of the Congo (which is experiencing its own, separate outbreak of Ebola right now), Uganda, and Sudan. None of the affected countries in the current outbreak had ever seen a case of Ebola; now, they’re the epicenter of the worst outbreak in history.
Ebola’s emergence in the major West African cities of Conakry (Guinea), Freetown (Sierra Leone), Monrovia (Liberia), and Lagos (Nigeria) has also been unique to the current outbreak. Never before has Ebola affected such large population centers, nor has it ever spread across such a large swath of land.
And more than 8 months into the outbreak, the virus’ reach is expanding exponentially, with more cases reported in the past week than in any other thus far. The doubling period for the virus — the length of time it takes for the cases to double in number — is just 34.8 days.
These unusual (and alarming) characteristics have left scientists scrambling to answer two key questions: Where did the virus come from? And has the strain changed since it first emerged?
Figuring out the answer could yield important information about the spread of Ebola — from animal-to-human and from human-to-human — that may help public health officials better prepare for, and possibly even prevent, future outbreaks. It could also have important implications for scientists as they work to develop an effective treatment for the virus.
With that in mind, an international team of researchers set out on the task of sequencing nearly 100 Ebola genomes collected during the 2014 outbreak.

Uncovering the genetic origins of the current Ebola outbreak could help stop the spread of the virus and even help public health officials prepare for future outbreaks.
“In an ongoing public health crisis, where accurate and timely information is crucial, new genomic technologies can provide near real-time insights into the pathogen’s origin, transmission dynamics, and evolution,” the researchers write. “We used massively parallel viral sequencing to understand how and when [Ebola] entered human populations in the 2014 West African outbreak, whether the outbreak is continuing to be fed by new transmissions from its natural reservoir, and how the virus changed, both before and after its recent jump to humans.
The research team, including members from the Broad Institute and Harvard University in the US and the Sierra Leone Ministry of Health and Sanitation, has seen firsthand just how deadly the virus is: Five team members — including Dr. Sheik Humarr Khan, the beloved “hero doctor” from the Kenema Government Hospital — died of Ebola before the manuscript was published. Their fellow authors honor their memory in the study report.
The team carried out their analysis on 99 genomes that came from 78 different patients diagnosed with Ebola virus disease in Sierra Leone during the first 24 days of the outbreak. Importantly, some patients gave more than one sample, allowing the team to see how the virus changed over the course of a single infection.
Using deep sequencing techniques, the team increased the amount of data available on the Ebola virus by four-fold. The results show that the 2014 Ebola virus genomes contain over 300 mutations that distinguish them from previous outbreaks. The team also found clues to suggest the 2014 outbreak started from a single introduction into humans and then spread among them over many months.

Genomic sequencing of nearly 100 Ebola genomes from the outbreak in Sierra Leone yielded key information about where the disease came from and how it spread.
Taking a more in-depth look at the results, the study yielded three key findings.
Emergence in Sierra Leone
Firstly, the analysis showed that Ebola’s arrival in Sierra Leone in May likely started with a funeral. A young pregnant woman tested positive for the virus and was treated at Kenema Government Hospital. Health workers who traced her contacts discovered that she and more than a dozen other women recently had attended the burial of a traditional healer who had been treating Ebola patients near the Sierra Leone-Guinea border. All of them had been infected.
“They realized she was not an isolated case,” says co-senior author Dr. Pardis Sabeti, associate professor at Harvard University and a senior associate member at the Broad Institute. Dr. Sabeti’s lab sequenced the Ebola genomes and quickly made public the data earlier this summer.
Common origins with 1976 strain
Secondly, the genomic sequencing also offers hints as to how the Ebola “Zaire” strain at the heart of the current outbreak — one of five types of Ebola virus known to infect humans — likely ended up in West Africa in the first place. Researchers said the data suggests that the virus spread from an animal host, possibly bats, and that diverged around 2004 from an Ebola strain in central Africa. Fruit bats are thought to be the natural host of the Ebola virus, which first appeared among humans in simultaneous outbreaks in Nzara in Sudan, and in Yambuku in the Democratic Republic of the Congo (DRC), in 1976.
“We don’t actually know where the virus has been since then,” says Dr. Sabeti, referring to the time between 2004 (when the strain diverged) and when the virus resurfaced earlier this year. “We’re trying to piece together an historical record.”
Rapidly mutating virus
Thirdly, Thursday’s study also details hundreds of genetic mutations that make the current Ebola outbreak different from any in the past. Some of those changes have the potential to affect the accuracy of diagnostic tests or the effectiveness of vaccines and treatments under development for the disease.
With their findings, the researchers presented a catalog of 395 mutations, of which over 340 distinguish the current outbreak from previous ones. Fifty of the mutations were acquired in the first month alone, the researchers found.
“We’ve uncovered more than 300 genetic clues about what sets this outbreak apart from previous outbreaks,” Stephen Gire, one of the study’s co-authors and an infectious disease researcher at Harvard, said in an announcement about the findings. “Although we don’t know whether these differences are related to the severity of the current outbreak, by sharing these data with the research community, we hope to speed up our understanding of this epidemic and support global efforts to contain it.”
Scientists say the findings also underscore the importance of getting the outbreak under control as quickly as possible, given that the risks associated with the virus rise as it continues to spread. As Dr. Charles Chiu, an infectious-disease physician at the University of California, San Francisco, explained to Nature News:
“The longer we allow the outbreak to continue, the greater the opportunity the virus has to mutate, and it’s possible that it will mutate into a form that would be an even greater threat than it is right now.”